SARS-CoV-2 Protein Homology Models
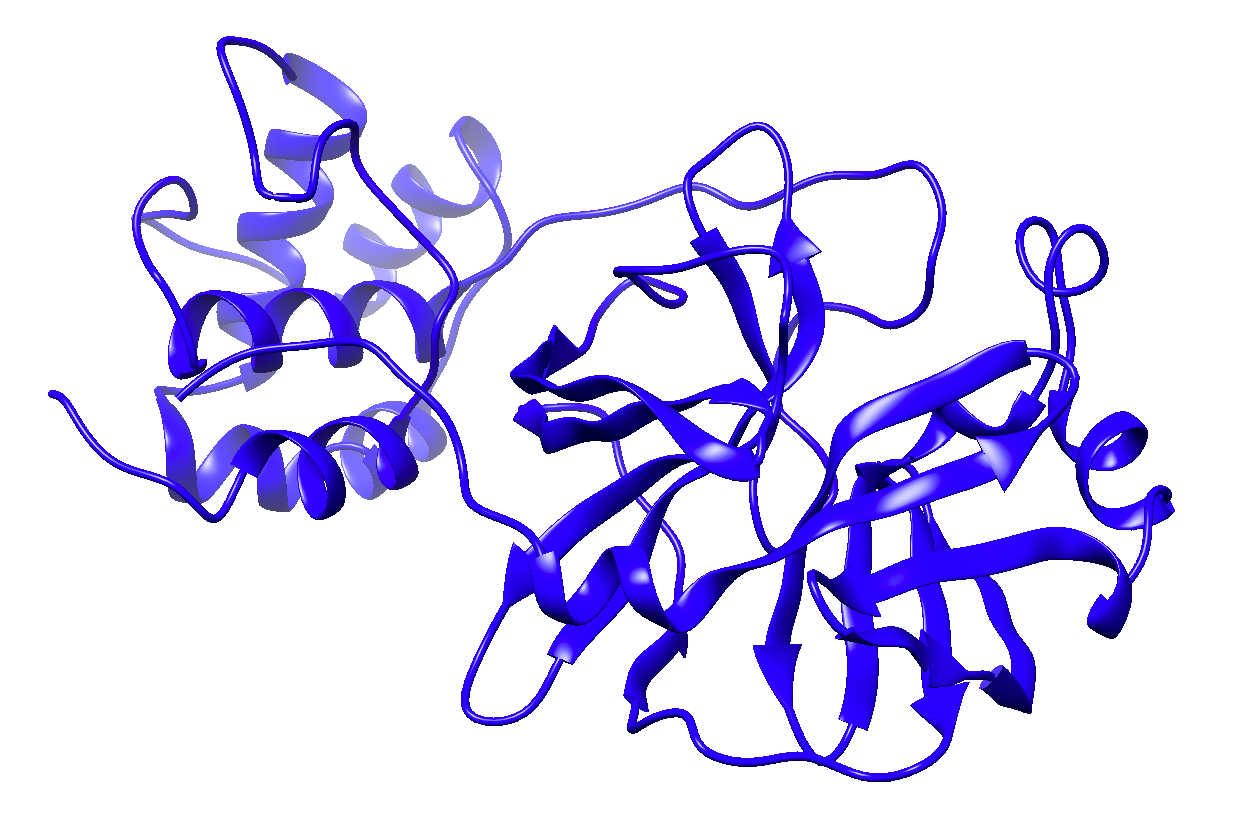
Models of the SARS-CoV-2 proteins – downloadable in PDB format on this page – were constructed using the AS2TS software package.
Protein sequences come from the SARS-CoV-2 reference genome here and here. Names and annotations in the following table come from that source.
The last part of each PDB file name indicates the model template by PDB ID and chain. For example, the nsp1 model is based on template PDB 2gdt, chain A. Each file includes additional information in REMARK fields.
|
Name |
Long name |
Annotation |
Protein |
Length |
Coverage |
Homology models |
||
|---|---|---|---|---|---|---|---|---|
|
nsp1 |
leader protein |
180 | 13-127 |
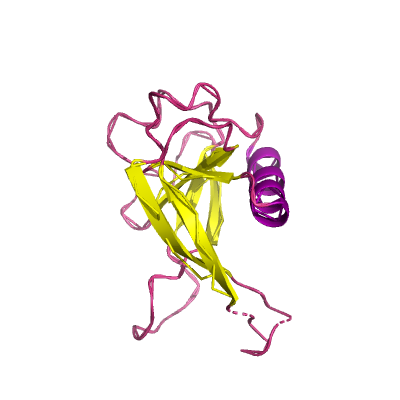 2gdt_A 2gdt_A
|
||||
|
nsp2 |
638 | |||||||
|
nsp3 |
conserved domains are: N-terminal acidic (Ac), predicted phosphoesterase, papain-like proteinase, Y-domain, transmembrane domain 1 (TM1), adenosine diphosphate-ribose 1''-phosphatase (ADRP); |
1945 | 1-111 207-373 413-676 746-1064 1089-1203 |
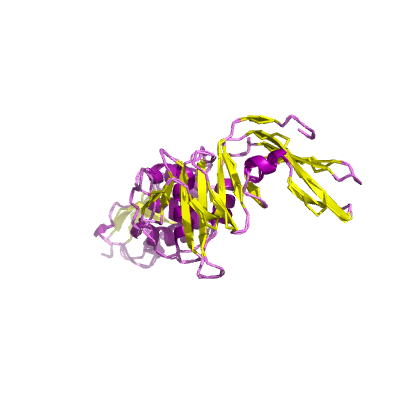 6w9c_A 6w9c_A
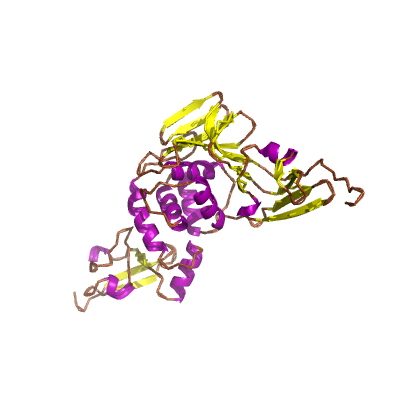 5e6j_A 5e6j_A
 5tl7_B 5tl7_B
|
 2w2g_A 2w2g_A
 6w02_B 6w02_B
|
 2k87_A 2k87_A
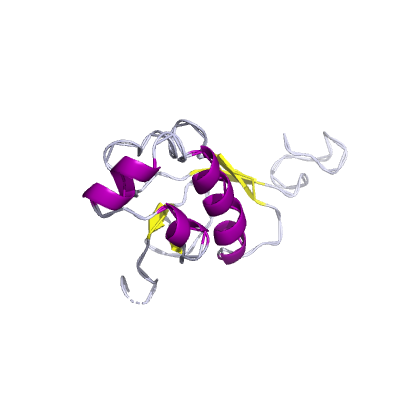 2gri_A 2gri_A
|
||
|
nsp4 |
nsp4B_TM |
contains transmembrane domain 2 (TM2); |
500 | 403-499 |
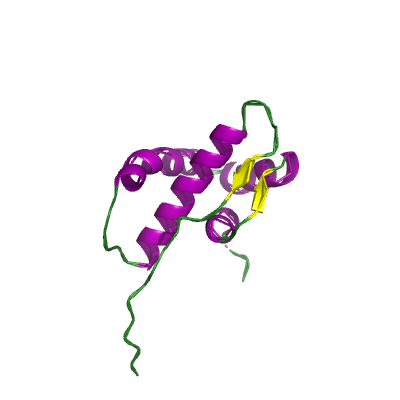 3vcb_A 3vcb_A
|
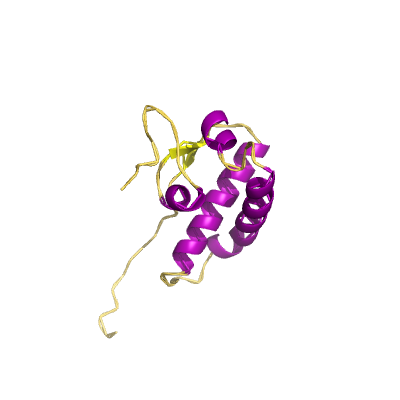 3gzf_A 3gzf_A
|
||
|
nsp5 |
3C-like proteinase |
nsp5A_3CLpro and nsp5B_3CLpro; main proteinase (Mpro); mediates cleavages downstream of nsp4. 3D structure of the SARSr-CoV homolog has been determined (Yang et al., 2003); |
306 | 1-306 |
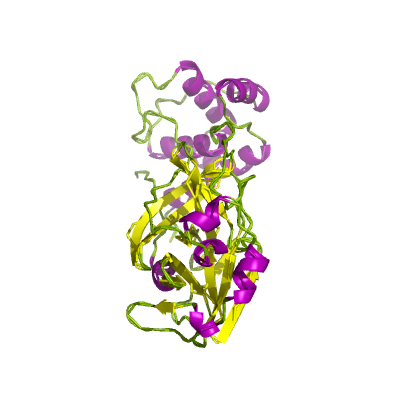 5r82_A 5r82_A
|
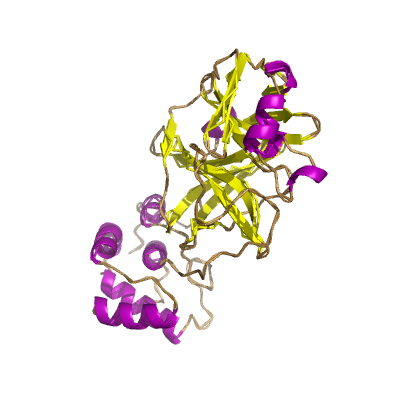 6lu7_A 6lu7_A
|
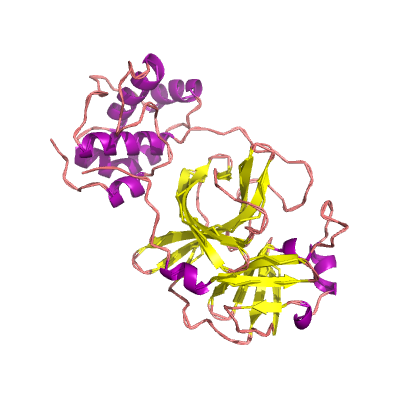 3tnt_A 3tnt_A
|
|
|
nsp6 |
nsp6_TM |
putative transmembrane domain |
290 | |||||
|
nsp7 |
83 | 1-83 |
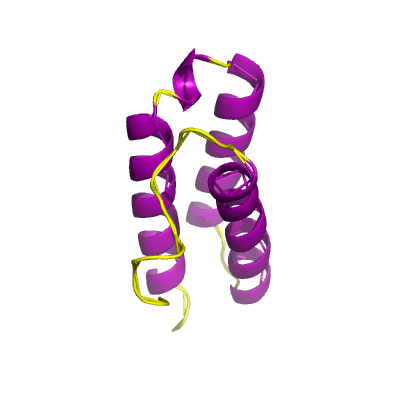 6m71_C 6m71_C
|
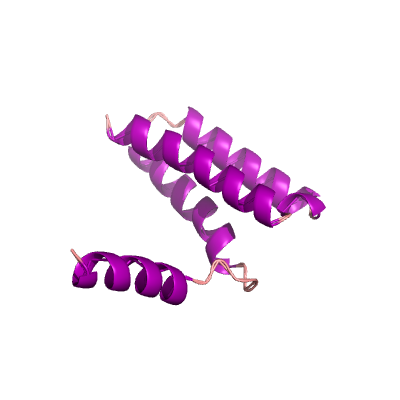 5f22_A 5f22_A
|
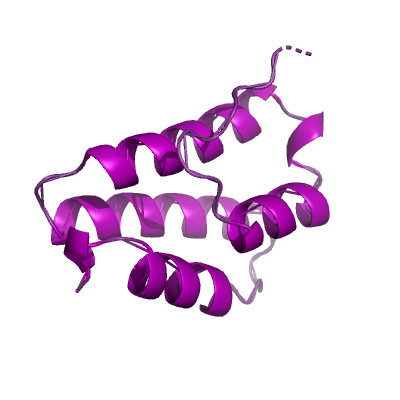 3ub0_B 3ub0_B
|
|||
|
nsp8 |
198 | 1-191 |
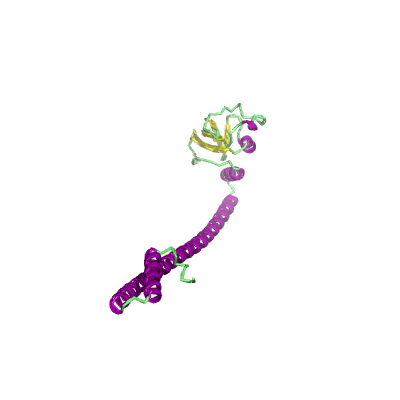 2ahm_G 2ahm_G
|
|||||
|
nsp9 |
ssRNA-binding protein; |
113 | 1-113 |
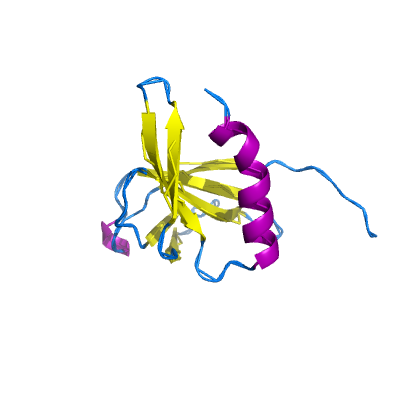 6w9q_A 6w9q_A
|
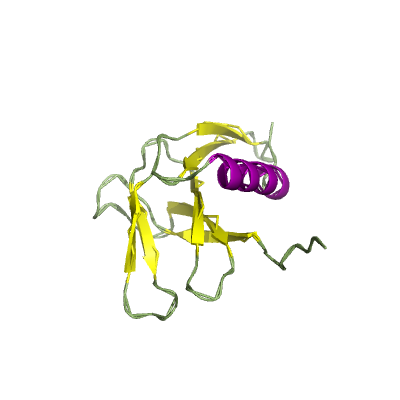 6w4b_B 6w4b_B
|
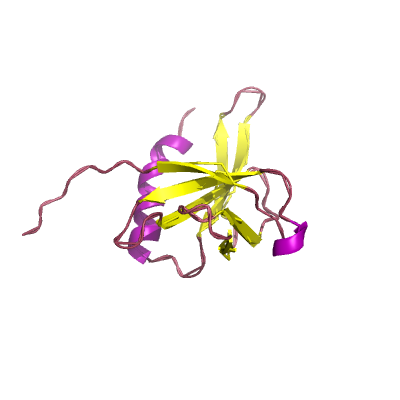 1uw7_A 1uw7_A
|
||
|
nsp10 |
nsp10_CysHis |
formerly known as growth-factor-like protein (GFL); |
139 | 1-131 |
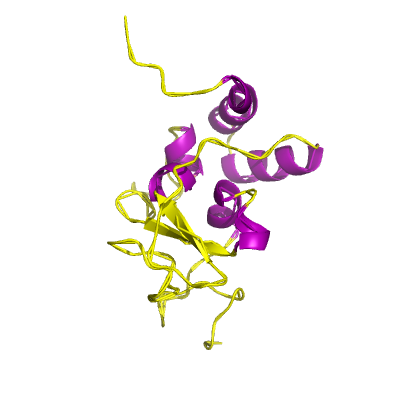 5c8t_A 5c8t_A
|
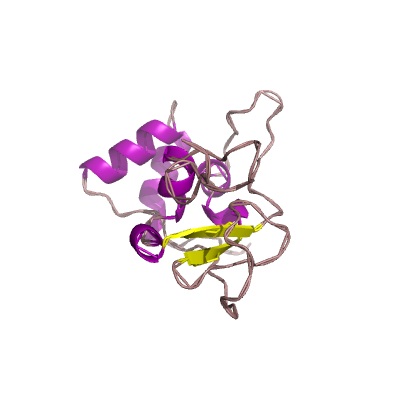 5nfy_N 5nfy_N
|
||
|
nsp12 |
RNA-dependent RNA polymerase |
NiRAN and RdRp |
932 | 1-932 |
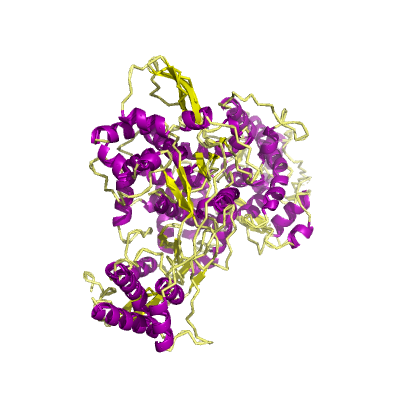 6m71_A 6m71_A
|
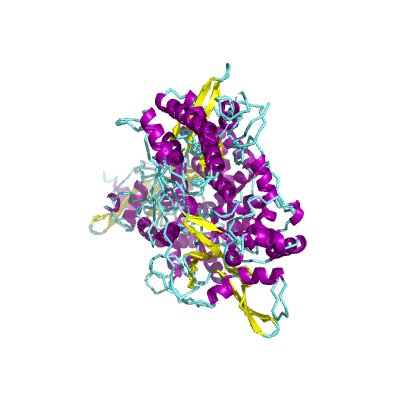 7btf_A 7btf_A
|
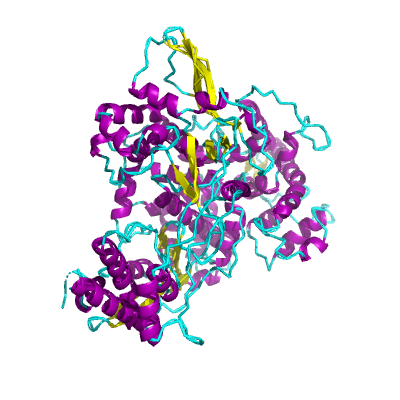 6nur_A 6nur_A
|
|
|
nsp13 |
helicase |
nsp13_ZBD, nsp13_TB, and nsp_HEL1core; zinc-binding domain (ZD), NTPase/helicase domain (HEL), RNA 5'-triphosphatase |
601 | 1-596 |
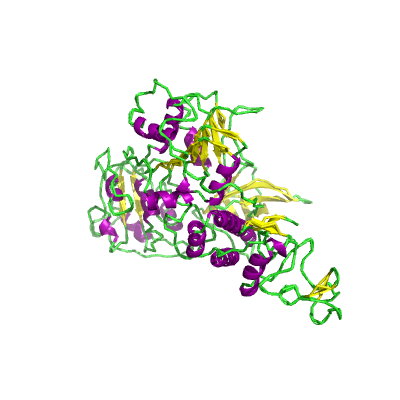 6jyt_A 6jyt_A
|
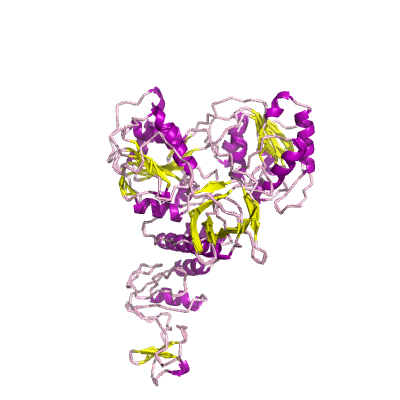 5wwp_B 5wwp_B
|
||
|
nsp14 |
3'-to-5' exonuclease" |
nsp14A2_ExoN and nsp14B_NMT; |
527 | 1-525 |
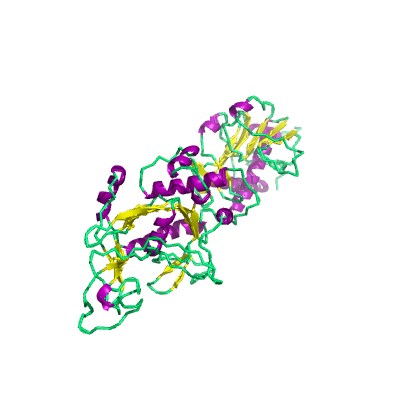 5c8s_B 5c8s_B
|
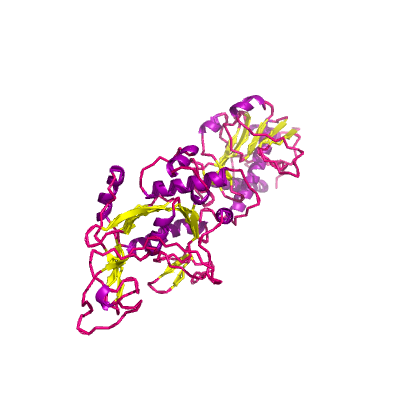 5c8t_B 5c8t_B
|
||
|
nsp15 |
endoRNAse |
nsp15-A1 and nsp15B-NendoU; |
346 | 1-345 |
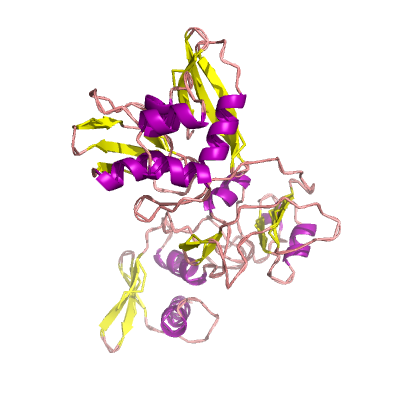 6w01_A 6w01_A
|
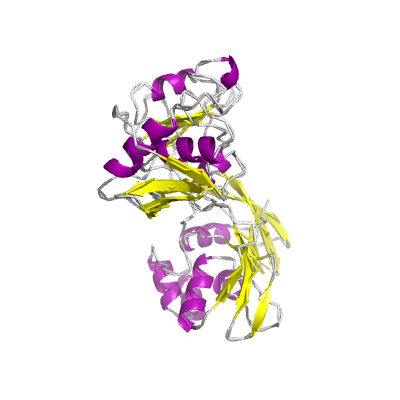 2h85_A 2h85_A
|
||
|
nsp16 |
2'-O-ribose methyltransferase |
nsp16_OMT; 2'-o-MT; |
298 | 1-298 |
 6w4h_A 6w4h_A
|
 6w61_A 6w61_A
|
||
|
S |
spike |
surface glycoprotein |
1273 | 18-1147 |
 6vsb_C 6vsb_C
 6vxx_B 6vxx_B
|
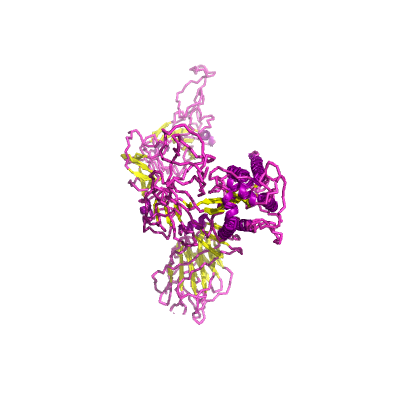 6acc_A 6acc_A
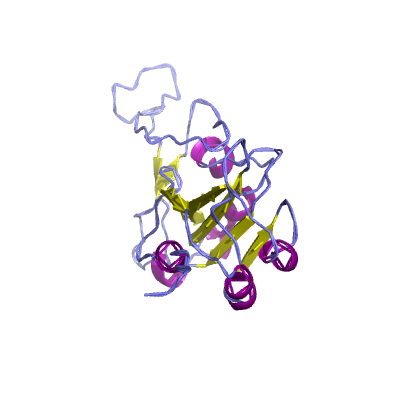 6m0j_E 6m0j_E
|
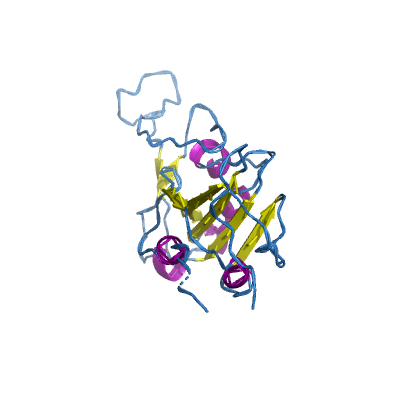 6lzg_B 6lzg_B
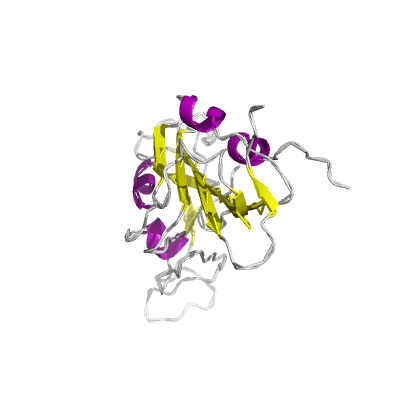 6w41_C 6w41_C
|
|
|
ORF3a |
ORF3a protein |
275 | ||||||
|
E |
envelope protein |
ORF4; structural protein; E protein |
75 |
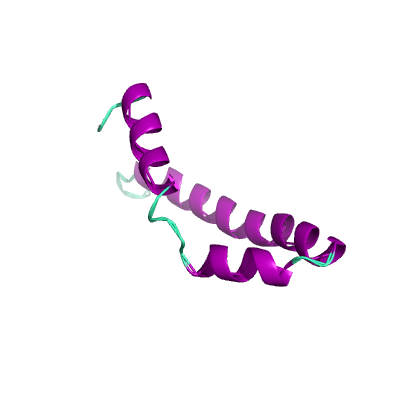 5x29_A 5x29_A
|
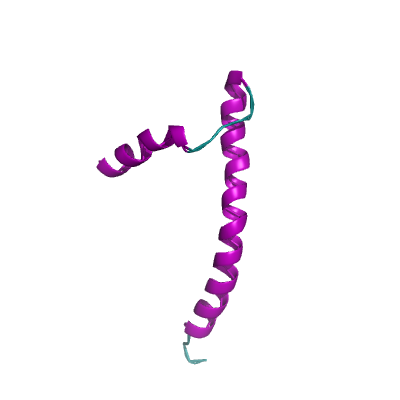 2mm4_A 2mm4_A
|
|||
|
M |
membrane glycoprotein |
ORF5; structural protein |
222 | |||||
|
ORF6 |
ORF6 protein |
61 | ||||||
|
ORF7a |
ORF7a protein |
121 | 15-98 |
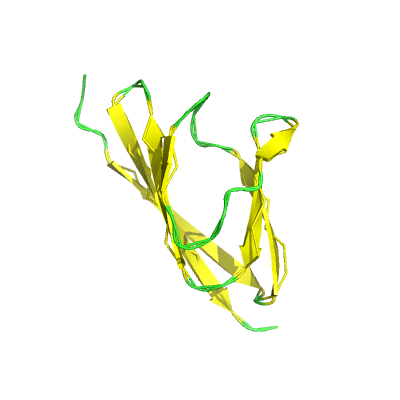 1xak_A 1xak_A
|
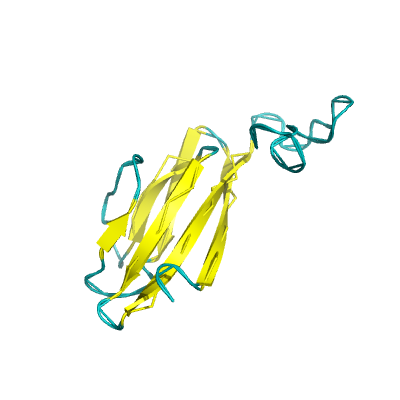 1yo4_A 1yo4_A
|
|||
|
ORF7b |
43 | |||||||
|
ORF8 |
ORF8 protein |
121 | ||||||
|
N |
nucleocapsid phosphoprotein |
ORF9; structural protein |
419 | 47-173 250-364 |
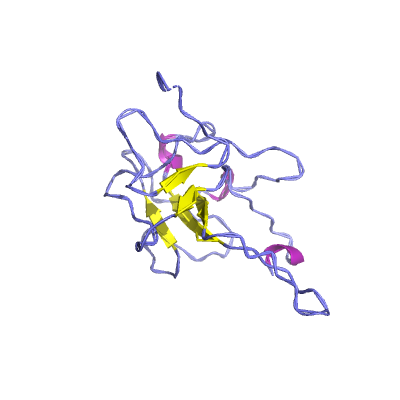 6yi3_A 6yi3_A
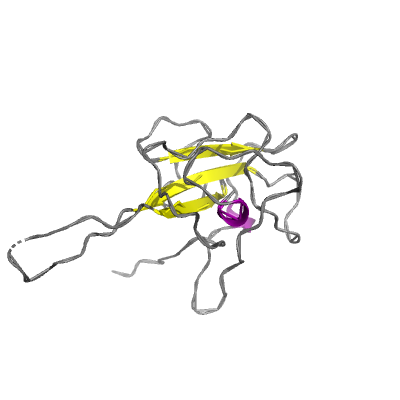 6m3m_B 6m3m_B
|
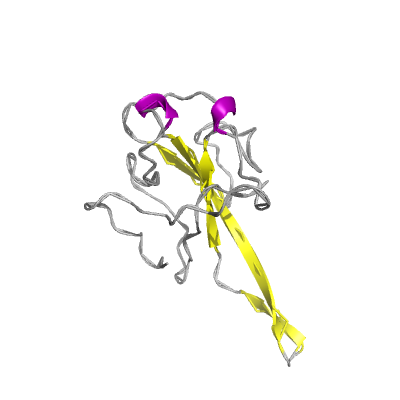 6vyo_D 6vyo_D
|
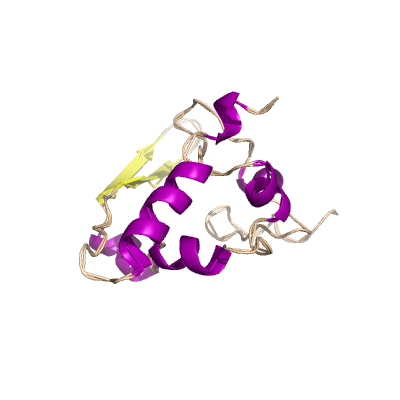 2cjr_A 2cjr_A
|
|
|
ORF10 |
ORF10 protein |
38 |